Reading Raster Data
RasterFrames registers a DataSource named raster that enables reading of GeoTIFFs (and other formats when GDAL is installed) from arbitrary URIs. The raster DataSource operates on either a single raster file location or another DataFrame, called a catalog, containing pointers to many raster file locations.
RasterFrames can also read from GeoTrellis catalogs and layers.
Single Rasters
The simplest way to use the raster reader is with a single raster from a single URI or file. In the examples that follow we’ll be reading from a Sentinel-2 scene stored in an AWS S3 bucket.
rf = spark.read.raster('https://rasterframes.s3.amazonaws.com/samples/luray_snp/B02.tif')
rf.printSchema()
root
|-- proj_raster_path: string (nullable = false)
|-- proj_raster: struct (nullable = true)
| |-- tile_context: struct (nullable = true)
| | |-- extent: struct (nullable = false)
| | | |-- xmin: double (nullable = false)
| | | |-- ymin: double (nullable = false)
| | | |-- xmax: double (nullable = false)
| | | |-- ymax: double (nullable = false)
| | |-- crs: struct (nullable = false)
| | | |-- crsProj4: string (nullable = false)
| |-- tile: tile (nullable = false)
The file at the address above is a valid Cloud Optimized GeoTIFF (COG), which RasterFrames fully supports. RasterFrames will take advantage of the optimizations in the COG format to enable more efficient reading compared to non-COG GeoTIFFs.
Let’s unpack the proj_raster column and look at the contents in more detail. It contains a CRS, a spatial extent measured in that CRS, and a two-dimensional array of numeric values called a tile.
crs = rf.select(rf_crs("proj_raster").alias("value")).first()
print("CRS", crs.value.crsProj4)
CRS +proj=utm +zone=17 +datum=WGS84 +units=m +no_defs
rf.select(
rf_extent("proj_raster").alias("extent"),
rf_tile("proj_raster").alias("tile")
)
Showing only top 5 rows.
| extent | tile |
|---|---|
| [807480.0, 4207860.0, 809760.0, 4223220.0] | |
| [792120.0, 4269300.0, 807480.0, 4284660.0] | |
| [699960.0, 4223220.0, 715320.0, 4238580.0] | |
| [807480.0, 4190220.0, 809760.0, 4192500.0] | |
| [715320.0, 4223220.0, 730680.0, 4238580.0] |
You can also see that the single raster has been broken out into many rows containing arbitrary non-overlapping regions. Doing so takes advantage of parallel in-memory reads from the cloud hosted data source and allows Spark to work on manageable amounts of data per row. The map below shows downsampled imagery with the bounds of the individual tiles.
The image contains visible “seams” between the tile extents due to reprojection and downsampling used to create the image. The native imagery in the DataFrame does not contain any gaps in the source raster’s coverage.
Let’s select a single tile and view it. The tile preview image as well as the string representation provide some basic information about the tile: its dimensions as numbers of columns and rows and the cell type, or data type of all the cells in the tile. For more about cell types, refer to this discussion.
tile = rf.select(rf_tile("proj_raster")).first()[0]
display(tile)
Multiple Singleband Rasters
In this example, we show the reading two bands of Landsat 8 imagery (red and near-infrared), combining them with rf_normalized_difference to compute NDVI, a common measure of vegetation health. As described in the section on catalogs, image URIs in a single row are assumed to be from the same scene/granule, and therefore compatible. This pattern is commonly used when multiple bands are stored in separate files.
bands = [f'B{b}' for b in [4, 5]]
uris = [f'https://landsat-pds.s3.us-west-2.amazonaws.com/c1/L8/014/032/LC08_L1TP_014032_20190720_20190731_01_T1/LC08_L1TP_014032_20190720_20190731_01_T1_{b}.TIF' for b in bands]
catalog = ','.join(bands) + '\n' + ','.join(uris)
rf = (spark.read.raster(catalog, bands)
# Adding semantic names
.withColumnRenamed('B4', 'red').withColumnRenamed('B5', 'NIR')
# Adding tile center point for reference
.withColumn('longitude_latitude', st_reproject(st_centroid(rf_geometry('red')), rf_crs('red'), lit('EPSG:4326')))
# Compute NDVI
.withColumn('NDVI', rf_normalized_difference('NIR', 'red'))
# For the purposes of inspection, filter out rows where there's not much vegetation
.where(rf_tile_sum('NDVI') > 10000)
# Order output
.select('longitude_latitude', 'red', 'NIR', 'NDVI'))
display(rf)
Showing only top 5 rows.
| longitude_latitude | red | NIR | NDVI |
|---|---|---|---|
| POINT (-74.54798498452405 40.38740603406… | |||
| POINT (-74.27356358544107 40.66276020383… | |||
| POINT (-74.64174679709124 39.76498729658… | |||
| POINT (-75.08989515249583 39.62710756189… | |||
| POINT (-73.99582313850216 41.00659730217… |
Multiband Rasters
A multiband raster is represented by a three dimensional numeric array stored in a single file. The first two dimensions are spatial, and the third dimension is typically designated for different spectral bands. The bands could represent intensity of different wavelengths of light (or other electromagnetic radiation), or they could measure other phenomena such as time, quality indications, or additional gas concentrations, etc.
Multiband rasters files have a strictly ordered set of bands, which are typically indexed from 1. Some files have metadata tags associated with each band. Some files have a color interpetation metadata tag indicating how to interpret the bands.
When reading a multiband raster or a catalog describing multiband rasters, you will need to know ahead of time which bands you want to read. You will specify the bands to read, indexed from zero, as a list of integers into the band_indexes parameter of the raster reader.
For example, we can read a four-band (red, green, blue, and near-infrared) image as follows. The individual rows of the resulting DataFrame still represent distinct spatial extents, with a projected raster column for each band specified by band_indexes.
mb = spark.read.raster(
'https://rasterframes.s3.amazonaws.com/samples/naip/m_3807863_nw_17_1_20160620.tif',
band_indexes=[0, 1, 2, 3],
)
display(mb)
Showing only top 5 rows.
If a band is passed into band_indexes that exceeds the number of bands in the raster, a projected raster column will still be generated in the schema but the column will be full of null values.
You can also pass a catalog and band_indexes together into the raster reader. This will create a projected raster column for the combination of all items in catalog_col_names and band_indexes. Again if a band in band_indexes exceeds the number of bands in a raster, it will have a null value for the corresponding column.
Here is a trivial example with a catalog over multiband rasters. We specify two columns containing URIs and two bands, resulting in four projected raster columns.
import pandas as pd
mb_cat = pd.DataFrame([
{'foo': 'https://rasterframes.s3.amazonaws.com/samples/naip/m_3807863_nw_17_1_20160620.tif',
'bar': 'https://rasterframes.s3.amazonaws.com/samples/naip/m_3807863_nw_17_1_20160620.tif'
},
])
mb2 = spark.read.raster(
spark.createDataFrame(mb_cat),
catalog_col_names=['foo', 'bar'],
band_indexes=[0, 1],
tile_dimensions=(64,64)
)
mb2.printSchema()
root
|-- foo_path: string (nullable = false)
|-- bar_path: string (nullable = false)
|-- foo_b0: struct (nullable = true)
| |-- tile_context: struct (nullable = true)
| | |-- extent: struct (nullable = false)
| | | |-- xmin: double (nullable = false)
| | | |-- ymin: double (nullable = false)
| | | |-- xmax: double (nullable = false)
| | | |-- ymax: double (nullable = false)
| | |-- crs: struct (nullable = false)
| | | |-- crsProj4: string (nullable = false)
| |-- tile: tile (nullable = false)
|-- foo_b1: struct (nullable = true)
| |-- tile_context: struct (nullable = true)
| | |-- extent: struct (nullable = false)
| | | |-- xmin: double (nullable = false)
| | | |-- ymin: double (nullable = false)
| | | |-- xmax: double (nullable = false)
| | | |-- ymax: double (nullable = false)
| | |-- crs: struct (nullable = false)
| | | |-- crsProj4: string (nullable = false)
| |-- tile: tile (nullable = false)
|-- bar_b0: struct (nullable = true)
| |-- tile_context: struct (nullable = true)
| | |-- extent: struct (nullable = false)
| | | |-- xmin: double (nullable = false)
| | | |-- ymin: double (nullable = false)
| | | |-- xmax: double (nullable = false)
| | | |-- ymax: double (nullable = false)
| | |-- crs: struct (nullable = false)
| | | |-- crsProj4: string (nullable = false)
| |-- tile: tile (nullable = false)
|-- bar_b1: struct (nullable = true)
| |-- tile_context: struct (nullable = true)
| | |-- extent: struct (nullable = false)
| | | |-- xmin: double (nullable = false)
| | | |-- ymin: double (nullable = false)
| | | |-- xmax: double (nullable = false)
| | | |-- ymax: double (nullable = false)
| | |-- crs: struct (nullable = false)
| | | |-- crsProj4: string (nullable = false)
| |-- tile: tile (nullable = false)
URI Formats
RasterFrames relies on three different I/O drivers, selected based on a combination of scheme, file extentions, and library availability. GDAL is used by default if a compatible version of GDAL (>= 2.4) is installed, and if GDAL supports the specified scheme. If GDAL is not available, either the Java I/O or Hadoop driver will be selected, depending on scheme.
Note: The GDAL driver is the only one that can read non-GeoTIFF files.
| Prefix | GDAL | Java I/O | Hadoop |
|---|---|---|---|
gdal://<vsidrv>// |
yes | no | no |
file:// |
yes | yes | no |
http:// |
yes | yes | no |
https:// |
yes | yes | no |
ftp:// |
/vsicurl/ |
yes | no |
hdfs:// |
/vsihdfs/ |
no | yes |
s3:// |
/vsis3/ |
yes | no |
s3n:// |
no | no | yes |
s3a:// |
no | no | yes |
wasb:// |
/vsiaz/ |
no | yes |
wasbs:// |
no | no | yes |
Specific GDAL Virtual File System drivers can be selected using the gdal://<vsidrv>// syntax. For example If you have a archive.zip file containing a GeoTiff named my-file-inside.tif, you can address it with gdal://vsizip//path/to/archive.zip/my-file-inside.tif. Another example would be a MRF file in an S3 bucket on AWS: gdal://vsis3/my-bucket/prefix/to/raster.mrf. See the GDAL documentation for the format of the URIs after the gdal:/ scheme.
Raster Catalogs
Consider the definition of a Catalog previously discussed, let’s read the raster data contained in the catalog URIs. We will start with the external catalog of MODIS surface reflectance.
from pyspark import SparkFiles
from pyspark.sql import functions as F
cat_filename = "2018-07-04_scenes.txt"
spark.sparkContext.addFile("https://modis-pds.s3.amazonaws.com/MCD43A4.006/{}".format(cat_filename))
modis_catalog = spark.read \
.format("csv") \
.option("header", "true") \
.load(SparkFiles.get(cat_filename)) \
.withColumn('base_url',
F.concat(F.regexp_replace('download_url', 'index.html$', ''), 'gid')
) \
.drop('download_url') \
.withColumn('red' , F.concat('base_url', F.lit("_B01.TIF"))) \
.withColumn('nir' , F.concat('base_url', F.lit("_B02.TIF")))
print("Available scenes: ", modis_catalog.count())
Available scenes: 297
modis_catalog
Showing only top 5 rows.
MODIS data products are delivered on a regular, consistent grid, making identification of a specific area over time easy using (h,v) grid coordinates (see below).
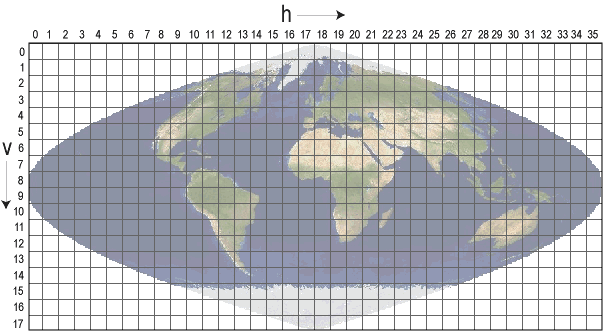
For example, MODIS data right above the equator is all grid coordinates with v07.
equator = modis_catalog.where(F.col('gid').like('%v07%'))
equator.select('date', 'gid')
Showing only top 5 rows.
| date | gid |
|---|---|
| 2018-07-04 00:00:00 | MCD43A4.A2018185.h07v07.006.2018194033213 |
| 2018-07-04 00:00:00 | MCD43A4.A2018185.h08v07.006.2018194033224 |
| 2018-07-04 00:00:00 | MCD43A4.A2018185.h09v07.006.2018194033547 |
| 2018-07-04 00:00:00 | MCD43A4.A2018185.h26v07.006.2018194033904 |
| 2018-07-04 00:00:00 | MCD43A4.A2018185.h12v07.006.2018194033912 |
Now that we have prepared our catalog, we simply pass the DataFrame or CSV string to the raster DataSource to load the imagery. The catalog_col_names parameter gives the columns that contain the URI’s to be read.
rf = spark.read.raster(equator, catalog_col_names=['red', 'nir'])
rf.printSchema()
root
|-- red_path: string (nullable = false)
|-- nir_path: string (nullable = false)
|-- red: struct (nullable = true)
| |-- tile_context: struct (nullable = true)
| | |-- extent: struct (nullable = false)
| | | |-- xmin: double (nullable = false)
| | | |-- ymin: double (nullable = false)
| | | |-- xmax: double (nullable = false)
| | | |-- ymax: double (nullable = false)
| | |-- crs: struct (nullable = false)
| | | |-- crsProj4: string (nullable = false)
| |-- tile: tile (nullable = false)
|-- nir: struct (nullable = true)
| |-- tile_context: struct (nullable = true)
| | |-- extent: struct (nullable = false)
| | | |-- xmin: double (nullable = false)
| | | |-- ymin: double (nullable = false)
| | | |-- xmax: double (nullable = false)
| | | |-- ymax: double (nullable = false)
| | |-- crs: struct (nullable = false)
| | | |-- crsProj4: string (nullable = false)
| |-- tile: tile (nullable = false)
|-- date: string (nullable = true)
|-- gid: string (nullable = true)
|-- base_url: string (nullable = true)
Observe the schema of the resulting DataFrame has a projected raster struct for each column passed in catalog_col_names. For reference, the URI is now in a column appended with _path. Taking a quick look at the representation of the data, we see again each row contains an arbitrary portion of the entire scene coverage. We also see that for two-D catalogs, each row contains the same spatial extent for all tiles in that row.
sample = rf \
.select('gid', rf_extent('red'), rf_extent('nir'), rf_tile('red'), rf_tile('nir')) \
.where(~rf_is_no_data_tile('red'))
sample.limit(3)
| gid | rf_extent(red) | rf_extent(nir) | rf_tile(red) | rf_tile(nir) |
|---|---|---|---|---|
| MCD43A4.A2018185.h16v07.006.2018194033835 | [-1156428.54045364, 1630860.7621777998, -1111950.519667, 1749468.8176088398] | [-1156428.54045364, 1630860.7621777998, -1111950.519667, 1749468.8176088398] | ||
| MCD43A4.A2018185.h22v07.006.2018194034134 | [4566410.13409804, 1111950.519667, 4685018.18952908, 1156428.54045364] | [4566410.13409804, 1111950.519667, 4685018.18952908, 1156428.54045364] | ||
| MCD43A4.A2018185.h33v07.006.2018194035136 | [1.7628122238449175E7, 1275036.59588468, 1.774673029388032E7, 1393644.65131572] | [1.7628122238449175E7, 1275036.59588468, 1.774673029388032E7, 1393644.65131572] |
Lazy Raster Reading
By default, reading raster pixel values is delayed until it is absolutely needed. The DataFrame will contain metadata and pointers to the appropriate portion of the data until reading of the source raster data is absolutely necessary. This can save a significant of computation and I/O time for two reasons. First, a catalog may contain a large number of rows. Second, the raster DataSource attempts to apply spatial preciates (e.g. where/WHERE clauses with st_intersects, et al.) at row creation, reducing the chance of unneeded data being fetched.
Consider the following two reads of the same data source. In the first, the lazy case, there is a pointer to the URI, extent and band to read. This will not be evaluated until the cell values are absolutely required. The second case shows the option to force the raster to be fully loaded right away.
uri = 'https://rasterframes.s3.amazonaws.com/samples/luray_snp/B02.tif'
lazy = spark.read.raster(uri).select(col('proj_raster.tile').cast('string'))
lazy
Showing only top 5 rows.
| tile |
|---|
| RasterRefTile(RasterRef(JVMGeoTiffRasterSource(https://rasterframes.s3.amazonaws.com/samples/luray_snp/B02.tif),0,Some(Extent(807480.0, 4207860.0, 809760.0, 4223220.0)),Some(GridBounds(1792,1280,1829,1535)))) |
| RasterRefTile(RasterRef(JVMGeoTiffRasterSource(https://rasterframes.s3.amazonaws.com/samples/luray_snp/B02.tif),0,Some(Extent(792120.0, 4269300.0, 807480.0, 4284660.0)),Some(GridBounds(1536,256,1791,511)))) |
| RasterRefTile(RasterRef(JVMGeoTiffRasterSource(https://rasterframes.s3.amazonaws.com/samples/luray_snp/B02.tif),0,Some(Extent(699960.0, 4223220.0, 715320.0, 4238580.0)),Some(GridBounds(0,1024,255,1279)))) |
| RasterRefTile(RasterRef(JVMGeoTiffRasterSource(https://rasterframes.s3.amazonaws.com/samples/luray_snp/B02.tif),0,Some(Extent(807480.0, 4190220.0, 809760.0, 4192500.0)),Some(GridBounds(1792,1792,1829,1829)))) |
| RasterRefTile(RasterRef(JVMGeoTiffRasterSource(https://rasterframes.s3.amazonaws.com/samples/luray_snp/B02.tif),0,Some(Extent(715320.0, 4223220.0, 730680.0, 4238580.0)),Some(GridBounds(256,1024,511,1279)))) |
non_lazy = spark.read.raster(uri, lazy_tiles=False).select(col('proj_raster.tile').cast('string'))
non_lazy
Showing only top 5 rows.
| tile |
|---|
| ArrayTile(256,256,uint16raw) |
| ArrayTile(38,256,uint16raw) |
| ArrayTile(256,256,uint16raw) |
| ArrayTile(256,256,uint16raw) |
| ArrayTile(256,256,uint16raw) |
In the initial examples on this page, you may have noticed that the realized (non-lazy) tiles are shown, but we did not change lazy_tiles. Instead, we used rf_tile to explicitly request the realized tile from the lazy representation.
Spatial Indexing and Partitioning
This is an experimental feature, and may be removed.
It’s often desirable to take extra steps in ensuring your data is effectively distributed over your computing resources. One way of doing that is using something called a “space filling curve”, which turns an N-dimensional value into a one dimensional value, with properties that favor keeping entities near each other in N-space near each other in index space. In particular RasterFrames support space-filling curves mapping the geographic location of tiles to a one-dimensional index space called xz2. To have RasterFrames add a spatial index based partitioning on a raster reads, use the spatial_index_partitions parameter. By default it will use the same number of partitions as configured in spark.sql.shuffle.partitions.
df = spark.read.raster(uri, spatial_index_partitions=True)
df
Showing only top 5 rows.
| proj_raster_path | proj_raster | spatial_index |
|---|---|---|
| https://rasterframes.s3.amazonaws.com/samples/luray_snp/B02.tif | 55256634714 | |
| https://rasterframes.s3.amazonaws.com/samples/luray_snp/B02.tif | 55256634714 | |
| https://rasterframes.s3.amazonaws.com/samples/luray_snp/B02.tif | 55256634715 | |
| https://rasterframes.s3.amazonaws.com/samples/luray_snp/B02.tif | 55256656560 | |
| https://rasterframes.s3.amazonaws.com/samples/luray_snp/B02.tif | 55256722095 |
You can also pass a positive integer to the parameter to specify the number of desired partitions.
df = spark.read.raster(uri, spatial_index_partitions=800)
GeoTrellis
GeoTrellis Catalogs
GeoTrellis is one of the key libraries upon which RasterFrames is built. It provides a Scala language API for working with geospatial raster data. GeoTrellis defines a tile layer storage format for persisting imagery mosaics. RasterFrames provides a DataSource supporting both reading and writing GeoTrellis layers.
A GeoTrellis catalog is a set of GeoTrellis layers. We can read a DataFrame giving details of the content of a catalog using the syntax below. The scheme is typically hdfs or file, or a cloud storage provider like s3.
gt_cat = spark.read.geotrellis_catalog('scheme://path-to-gt-catalog')
GeoTrellis Layers
The catalog will give details on the individual layers available for query. We can read each layer with the URI to the catalog, the layer name, and the desired zoom level.
gt_layer = spark.read.geotrellis(path='scheme://path-to-gt-catalog', layer=layer_name, zoom=zoom_level)
This will return a RasterFrame with additional GeoTrellis-specific metadata, inherited from GeoTrellis, stored as JSON in the metadata of the tile column.